
Introduction: Barley is an important cereal grown mainly for feed and malting. You only mention one list so what you're trying to achieve is not clear.67th American Society for Mass Spectrometry Conference, Atlanta, Georgia, USA, June 2-6, 2019 The second one will show you the distribution of genes on the ontology.Īs for doing enrichment analysis, you need at least two lists to compare one against the other.

The first one will show the distribution of GO terms relative to the gene information you used to build the graph. However, note that the two options don't show the same information. As an alternative you could link the GO terms by some measure of semantic similarity. The ontology being a graph, you could just import it into Cytoscape from a compatible file format. Another option, which may be what you're asking about, would be to make a graph of GO terms and show the genes as attributes of the terms. What kind of information you consider to make a link is up to you depending on the question you're interested in. connect two genes in your data set if two known homologs can be linked. One option is to transfer this information by homology i.e. It seems your problem is that you don't have network information about your genes.
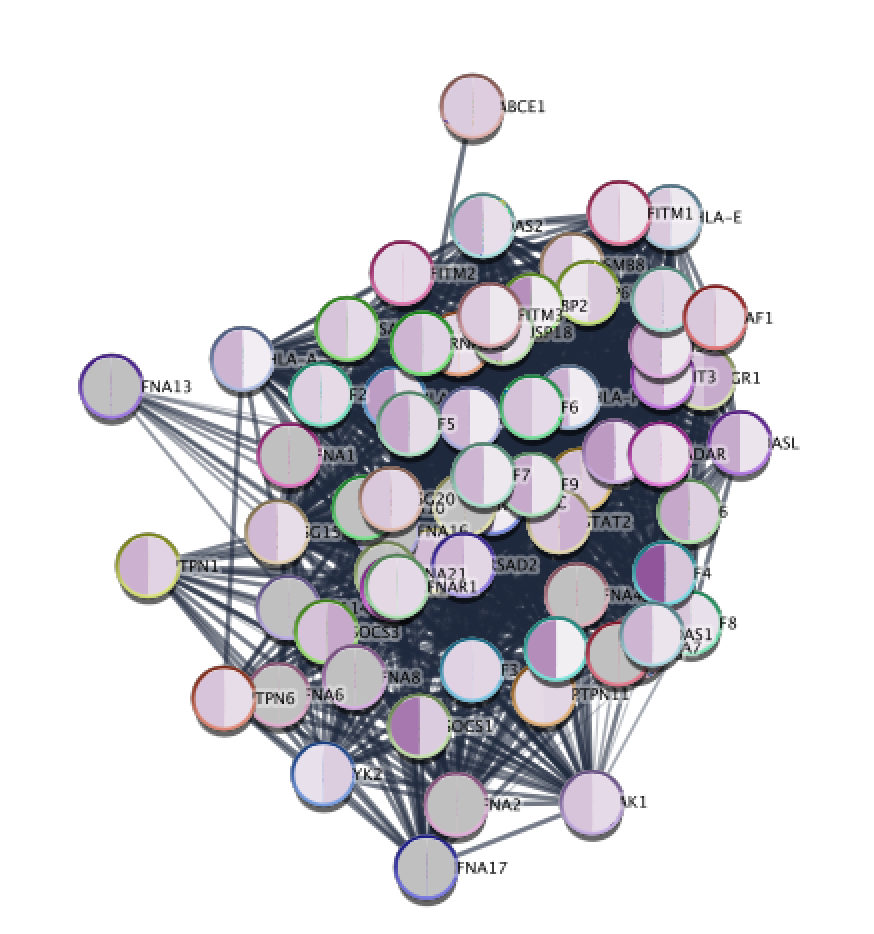
Just make the network you want/need with a script, save to a file and import into Cytoscape. OutLine = GOvalue + "\t" + splits + "\t" + splits + "\n"Įlif splits = "intersection_of:" or splits = "creat": import reīreak #read all the nodes, don't need type defs
Next step will be to create sub-graphs representing new input file GO terms (for multiple treatments), and then compare them. I honestly don't know the next step at this juncture. But the network needed to exist before I could do that. This is how I accomplished the task of making the GO network in Cytoscape.Ģ) Run my python script to convert go.obo to GO.cytoscape.sif fileģ) Cytoscape -> Import -> Network -> File, select the GO.cytoscape.sif fileĪs far as my goal not being clear, yes my end game is exactly to do a comparison / enrichment analysis. You would need to map the GO terms to create some metric of enrichment and association with GOSlims so that you can then import them to the Cytoscape to build the network associations and and then visualize. If this does not work you can always take a look at this thread and see if you can get some idea. This is what is coming to my mind as of now. Then you should be able to have for your GO term list enrichment score and p-value which can then be put to cytoscape to make the necessary relations for defining the edge and produce a network. But since you do not have gene lists so you directly want to do it with GO terms there needs to be some association score so that you can associate an enrichment score or association score of your GO terms with that of the entire GO master file that you have for metabolites. That is what will build the network, which usually one can make from gene lists putting them either in stringdb or with cytoscape with GO plugins and then build the relationships.

#Cytoscape and blast2go how to
I am just thinking of how to put the relations of GO terms to get the edges and connection for making the graph. In cytoscape there are some plugins like ClueGO and EnrichmentMap, but I have never used them for my purpose.


 0 kommentar(er)
0 kommentar(er)
